Enzyme
This plant enzyme, involved in the biosynthesis of anthocyanidins, is known to act on (+)-dihydrokaempferol, (+)-taxifolin, and (+)-dihydromyricetin, although some enzymes may act only on a subset of these compounds. Each dihydroflavonol is reduced to the corresponding cis-flavan-3,4-diol. NAD+ can act instead of NADP+, but more slowly.
DFR
Enzyme
Dihydroflavonol 4-reductase
Enzyme name
EC:1.1.1.219
Entry
Oxidoreductases;Acting on the CH-OH group of donors;With NAD+ or NADP+ as acceptor
Class
(2R,3S,4S)-leucoanthocyanidin:NADP+ 4-oxidoreductase
Sysname
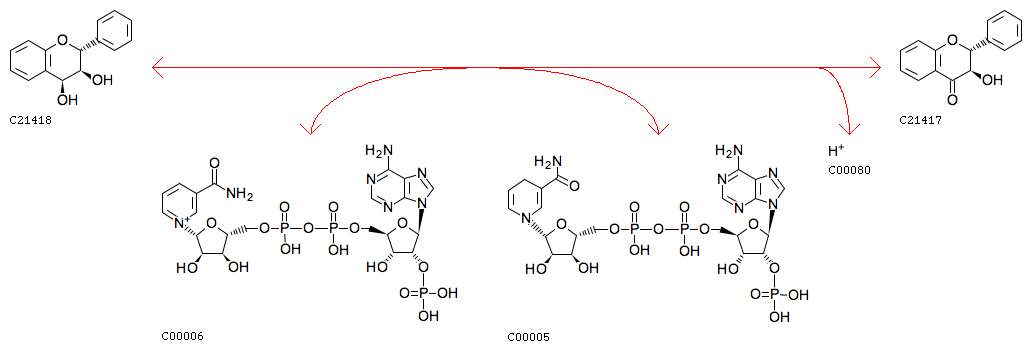
a (2R,3S,4S)-leucoanthocyanidin + NADP+ = a (2R,3R)-dihydroflavonol + NADPH + H+ [RN:R11500]
Reaction
(2R,3R)-dihydroflavonol [CPD:C21417];NADPH [CPD:C00005];H+ [CPD:C00080]
Product
Species information
Eremochloa ophiuroides
Species name
Eremochloa ophiuroides is a species of monocot in the family Poaceae (grass family).
Species info

Gene sequence info
Eremochloa_ophiuroides.DFR.fa
Download
>evm.model.ctg187.80
MQSSSPSRAFVEKELSYRGTILHDFSTFHFYVVVIPENRQSSGCFQKTKTDYVRSKTLSEKELLSYNDKQHQSGVEVVSLACALVGGDTIQAYLWRCWVEDACEAHVFCMEQASMAGRFLCAAGYPNMRDIVDHFAAKHPDLKIQLTEVTGERVRIVANTSKLEDLGFRFKYGVEEMLDCSVECGKRLGEL
>evm.model.ctg397.101
MGSVGVDAGGGGGSPDEEQQTEGSDGLPPPPAVCVTGSTGYVGSWLVRTLLRRGYRVHATARNTGKAWQVLGAVEEGSCRERLRVFRADMSEEGSFDAAVTGCVALFHVAASMDLHVSPALHQDNLEDHVMSSVLEPATRGTINVLQSCVRAGTVRRVVFTSSVSTLTAVDAAGRRKAVLDESCLRSLDDVWRTKPVGWIYILSKRLTEEAAFKFARENGVHLVSLVLPTVAGPFLTPSVPTSVQLLLSPVTGDPKLYAVLASVHARFGCVPLAHVQDACDAHVFLMEAPRAEGRYLCAAGGHAAAELARLLAERYPPFRPGGDRLSRDFDDASCSPPVVSSRRLLDLGFRFRYGVEDVVKDSVAQCLAHGFLEQT
>evm.model.ctg48.170
MEGAAIAKAGATKGPVLVTGASGYVGSWLVMKLLQAGYTVRATLRDPGNVAKTKPLLDLPGATERLSSWKADLAEEGSFDDAIRGCTGVFHVATPMDFESKDREVN
>evm.model.ctg48.171
MQGGRHRAPHRLHFLRRDVQHRGAAEARLRRGQLDRRRFLPSRQDDRMGTYVRSDFFACMATELTTTRFVYPYPCWQMYFVSKTLAEKAALAYAAEHGLDLISVIPTLVVGPFLSAGMPPSHITALALLTGNEAHYSILKQVQFVHLDDLCAAEIFLFEHPTAAAGRYVCSSHDATIHGLAAMLRERYPEFGIPQRFPGIEDDLQPVRFSSKKLQDLGFTFRCATVEDMYDAAIRTCREKGLIPLAATAGAGGDDGSAASVRAPGETDVTTTANGGGDCSASVRAPGEMDVTIGA
>evm.model.ctg48.168
MGEVVVAPRQAMEAGAGAGASVKGPVVVTGASGFVGSWLVMKLLQAGYTVRATVRDPANVAKTKPLLDLPGATERLSIWKADLAEEGSFDDAIRGCTGVFHVATPMDFESKDPENEVIKPTVEGMMSIMRACKEAGTVRRIVFTSSAGTVNIEERQRPVYDHDNWSDVEFCQRVKMTGWMYFVSKSLAEKAAMAYAAEHGLDLISVIPTLVVGPFLSAAMPPSLVTALALVTAGNEAHYSILKQVQFVHLDDLCDAEIFLFEHPAAAGRYVCSSHDATIHGLAAMLRDRYPQYDIPERFPGIQDDLQPVHFSSKKLLDHGFTFRYTVEDMFDAAIRTCREKGLIPLATPAEGTAHGSAPVCTPGERDVTIGA
MQSSSPSRAFVEKELSYRGTILHDFSTFHFYVVVIPENRQSSGCFQKTKTDYVRSKTLSEKELLSYNDKQHQSGVEVVSLACALVGGDTIQAYLWRCWVEDACEAHVFCMEQASMAGRFLCAAGYPNMRDIVDHFAAKHPDLKIQLTEVTGERVRIVANTSKLEDLGFRFKYGVEEMLDCSVECGKRLGEL
>evm.model.ctg397.101
MGSVGVDAGGGGGSPDEEQQTEGSDGLPPPPAVCVTGSTGYVGSWLVRTLLRRGYRVHATARNTGKAWQVLGAVEEGSCRERLRVFRADMSEEGSFDAAVTGCVALFHVAASMDLHVSPALHQDNLEDHVMSSVLEPATRGTINVLQSCVRAGTVRRVVFTSSVSTLTAVDAAGRRKAVLDESCLRSLDDVWRTKPVGWIYILSKRLTEEAAFKFARENGVHLVSLVLPTVAGPFLTPSVPTSVQLLLSPVTGDPKLYAVLASVHARFGCVPLAHVQDACDAHVFLMEAPRAEGRYLCAAGGHAAAELARLLAERYPPFRPGGDRLSRDFDDASCSPPVVSSRRLLDLGFRFRYGVEDVVKDSVAQCLAHGFLEQT
>evm.model.ctg48.170
MEGAAIAKAGATKGPVLVTGASGYVGSWLVMKLLQAGYTVRATLRDPGNVAKTKPLLDLPGATERLSSWKADLAEEGSFDDAIRGCTGVFHVATPMDFESKDREVN
>evm.model.ctg48.171
MQGGRHRAPHRLHFLRRDVQHRGAAEARLRRGQLDRRRFLPSRQDDRMGTYVRSDFFACMATELTTTRFVYPYPCWQMYFVSKTLAEKAALAYAAEHGLDLISVIPTLVVGPFLSAGMPPSHITALALLTGNEAHYSILKQVQFVHLDDLCAAEIFLFEHPTAAAGRYVCSSHDATIHGLAAMLRERYPEFGIPQRFPGIEDDLQPVRFSSKKLQDLGFTFRCATVEDMYDAAIRTCREKGLIPLAATAGAGGDDGSAASVRAPGETDVTTTANGGGDCSASVRAPGEMDVTIGA
>evm.model.ctg48.168
MGEVVVAPRQAMEAGAGAGASVKGPVVVTGASGFVGSWLVMKLLQAGYTVRATVRDPANVAKTKPLLDLPGATERLSIWKADLAEEGSFDDAIRGCTGVFHVATPMDFESKDPENEVIKPTVEGMMSIMRACKEAGTVRRIVFTSSAGTVNIEERQRPVYDHDNWSDVEFCQRVKMTGWMYFVSKSLAEKAAMAYAAEHGLDLISVIPTLVVGPFLSAAMPPSLVTALALVTAGNEAHYSILKQVQFVHLDDLCDAEIFLFEHPAAAGRYVCSSHDATIHGLAAMLRDRYPQYDIPERFPGIQDDLQPVHFSSKKLLDHGFTFRYTVEDMFDAAIRTCREKGLIPLATPAEGTAHGSAPVCTPGERDVTIGA
Gene tree
Gene tree

Sheng jun & Dong yang Teams, Yunnan agriculture University