Enzyme
Isolated from the plants Perilla frutescens var. crispa, Verbena hybrida [1], Dahlia variabilis [2] and Gentiana triflora (clustered gentian) [3]. It will also act on anthocyanidin 3-O-(6-O-malonylglucoside) [2] and is much less active with hydroxycinnamoylglucose derivatives [3]. There is no activity in the absence of the 3-O-glucoside group.
PF3R4
Enzyme
Anthocyanidin 3-O-glucoside 5-O-glucosyltransferase 1
Enzyme name
EC:2.4.1.298
Entry
Transferases;Glycosyltransferases;Hexosyltransferases
Class
UDP-alpha-D-glucose:anthocyanidin-3-O-beta-D-glucoside 5-O-glucosyltransferase
Sysname
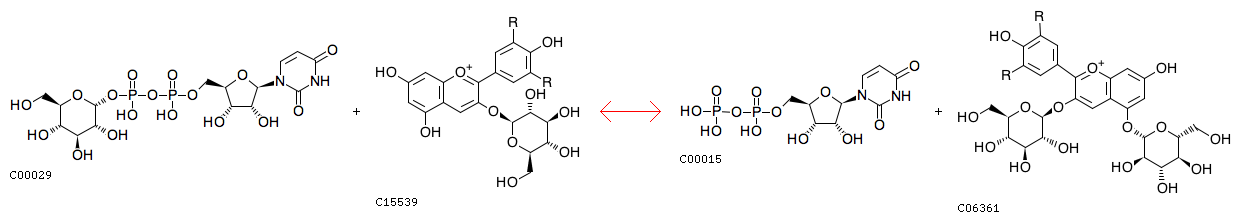
UDP-alpha-D-glucose + an anthocyanidin 3-O-beta-D-glucoside = UDP + an anthocyanidin 3,5-di-O-beta-D-glucoside [RN:R10262]
Reaction
UDP [CPD:C00015];anthocyanidin 3,5-di-O-beta-D-glucoside [CPD:C06361]
Product
Species information
Passiflora organensis
Species name
Passiflora organensis is a species of eudicot in the family Passifloraceae (passionflower famly).
Species info

Gene sequence info
Passiflora_organensis.PF3R4.fa
Download
>EVM%20prediction%20scaffold18_size2475805.88.mRNA1
MNPRFLLVTLPAQGHINPSLQCAKRLIRTGAHVTLVITVSVGRRMSRTSFPDGLSLAEFSDGFDDGFKSESDDHQRYMLEIKSRGSKALVDLIKGAENDGKPFTCLVYSMLLPWAAEVARVHGLPTACLWIQPATVLDIYYYYFNGYADIFSHGNDPSCRIELPGLPPLTARDLPSFLLSANLYAVILSMFEEQFEELNKEANPKMLVNTFEALEPEALRSIGDKFDLIGIGPLIPSAFLDGVDPSDTTFGCDIFHGGEDYTEWLNSKPISSVVYVSFGTIFVLSKPQMEAIARGLLACGRPFLWVIRAEQNEEEGAEKDNLLSYRQELEQQGKIVSWCSQVQLLSHSSIGCFMTHCGWNSTLETLASGIPVVAVPQWSDQRTNAKLIEDVWKTGVGVMKTNEEEIVEGEEIKRCLDLVMGGGELADQIRKNAKKWRDLARDAVKTGGSSDKNLKAFVDEFKLRMDMPLI
>EVM%20prediction%20scaffold18_size2475805.89.mRNA1
MARPAPNFLLLTFPAAGHIKPAFELAKRLIDIGASVTLAVCDSARPFIVETDIPKGLILFGFSDGYDNGVKPSDDLLLYMAEFKRRGTQAIVELLDARANDGIPISCILYTMLLPWVTEIARDKKVPSAFLWPQPATVLDIYYYYLNGYREAFDRCVDPSYVLELPGLPPFTSRDLPSLLGPSNVHTTTFLVLQEQLKKLYQETNTIILANTFDALEPEALKSIEYMNLIGIGPLNPDKSTSDKSKEAVQGTEDYINWLNTKPKASVVYASFGSLAILSDQQIEEITHGLLDTNRPFLWVIRTDQKLKEADIVSTRKDELRERGMIVPWCSQLEVLSHPSTGCFLSHCGWNSTMETLVHGIPVVGFPQWSDQRTNAKLIEDVWKTGVRVVPNEEGILGRDEIKRCLDLVMGDEEIAKQMNMNAKKFKDLARDAMKEGGSSDKNLRAFVEQVAQGRI
>EVM%20prediction%20scaffold37_size1024058.20.mRNA1
MVNLRILLVTFPAQGHINPALQFAKRLVALGAHVTFSTSIGAERRMDKTGASVPGLSFSAFHDGTEHSFRPTDDVDHYMFQLKHCGYNSLAQLIVASAEAGHPFTCVVYSNLIPWVGKVARDFHLPSFLLWNQSATVFDVFYHYFNGFEDAIKKSMSDPAFQLKLPGLLLPFASHDLPSFFSPTNRHIFGLKLMKEQLQILDQETNPPKVLVNTFDSLESEAIKSVGRYNLVGVGPLIPSAFLDGKDPSENSFGGDLFRASKDYMIWLDSQPQSSVIYISFGSVSVISNQEKEEMARALLDVGRPFLWVIRTGDAGGSEEKEDKPSCMEELERQGMIVPWCSQVEVLSHPSLGCFVTHCGWNSTLESLTSGVPVVAMPRWTDQLTNARLVNDISRTGVRVKSSSSAEKMFEAEEIKSCLDLVMGNEERGMEVRRNARKWKDLARETAKESGMSYKNLKAFVDEIAGGRDCT
>EVM%20prediction%20scaffold37_size1024058.21.mRNA1
MEREWRWNPGMFFFKAWTVSDISKTGVRVKASSSAERIFEAEEIKRCLDLVMGNDEGGMETRRNARIWKDLARETAKESGMSYKNLKAFVDEIAGG
>EVM%20prediction%20scaffold212_size72660.2.mRNA1
MVNLRILLVTFPAQGHINPALQFAKRLVALGAHVTFSTSFGAERMMDKTGASVPGLSFSAFHDGTEHSFRPTDDVDHYMFQLKHCGYNSLAQLIVASAEAGHPFTCVVYSNLIPWVGKVARDFHLPSFLLWNQSATVFDVFYHYFNGFEDAIKMSMSDPAFQLKLPGLSLSFASHDFPSFFSPTNRHTFGLKLMKEQLQILDQETNPPKVLVNTFDSLESEALKSVGRYNLVGVGPLIPSAFLDGKDPSENSFGGDLFRASKDYMIWLDSQPQSSVIYISFGSVSVISKQEKEEMARALLDVGRPFLWVIRTGDAGGSEEKEDKPSCMEELERQGMIVPWCSQVEVLSHPSLGCFVTHCGWNSTLESLTSGVPVVAMPRWTDQLTNARLVNDISRTGVRVKSSSSAEKMFEAEEIKSCLDLVMGNEERGMEVRRNARKWKDLARETAKESGMSYKNLKAFVDEIAGGRDCT
MNPRFLLVTLPAQGHINPSLQCAKRLIRTGAHVTLVITVSVGRRMSRTSFPDGLSLAEFSDGFDDGFKSESDDHQRYMLEIKSRGSKALVDLIKGAENDGKPFTCLVYSMLLPWAAEVARVHGLPTACLWIQPATVLDIYYYYFNGYADIFSHGNDPSCRIELPGLPPLTARDLPSFLLSANLYAVILSMFEEQFEELNKEANPKMLVNTFEALEPEALRSIGDKFDLIGIGPLIPSAFLDGVDPSDTTFGCDIFHGGEDYTEWLNSKPISSVVYVSFGTIFVLSKPQMEAIARGLLACGRPFLWVIRAEQNEEEGAEKDNLLSYRQELEQQGKIVSWCSQVQLLSHSSIGCFMTHCGWNSTLETLASGIPVVAVPQWSDQRTNAKLIEDVWKTGVGVMKTNEEEIVEGEEIKRCLDLVMGGGELADQIRKNAKKWRDLARDAVKTGGSSDKNLKAFVDEFKLRMDMPLI
>EVM%20prediction%20scaffold18_size2475805.89.mRNA1
MARPAPNFLLLTFPAAGHIKPAFELAKRLIDIGASVTLAVCDSARPFIVETDIPKGLILFGFSDGYDNGVKPSDDLLLYMAEFKRRGTQAIVELLDARANDGIPISCILYTMLLPWVTEIARDKKVPSAFLWPQPATVLDIYYYYLNGYREAFDRCVDPSYVLELPGLPPFTSRDLPSLLGPSNVHTTTFLVLQEQLKKLYQETNTIILANTFDALEPEALKSIEYMNLIGIGPLNPDKSTSDKSKEAVQGTEDYINWLNTKPKASVVYASFGSLAILSDQQIEEITHGLLDTNRPFLWVIRTDQKLKEADIVSTRKDELRERGMIVPWCSQLEVLSHPSTGCFLSHCGWNSTMETLVHGIPVVGFPQWSDQRTNAKLIEDVWKTGVRVVPNEEGILGRDEIKRCLDLVMGDEEIAKQMNMNAKKFKDLARDAMKEGGSSDKNLRAFVEQVAQGRI
>EVM%20prediction%20scaffold37_size1024058.20.mRNA1
MVNLRILLVTFPAQGHINPALQFAKRLVALGAHVTFSTSIGAERRMDKTGASVPGLSFSAFHDGTEHSFRPTDDVDHYMFQLKHCGYNSLAQLIVASAEAGHPFTCVVYSNLIPWVGKVARDFHLPSFLLWNQSATVFDVFYHYFNGFEDAIKKSMSDPAFQLKLPGLLLPFASHDLPSFFSPTNRHIFGLKLMKEQLQILDQETNPPKVLVNTFDSLESEAIKSVGRYNLVGVGPLIPSAFLDGKDPSENSFGGDLFRASKDYMIWLDSQPQSSVIYISFGSVSVISNQEKEEMARALLDVGRPFLWVIRTGDAGGSEEKEDKPSCMEELERQGMIVPWCSQVEVLSHPSLGCFVTHCGWNSTLESLTSGVPVVAMPRWTDQLTNARLVNDISRTGVRVKSSSSAEKMFEAEEIKSCLDLVMGNEERGMEVRRNARKWKDLARETAKESGMSYKNLKAFVDEIAGGRDCT
>EVM%20prediction%20scaffold37_size1024058.21.mRNA1
MEREWRWNPGMFFFKAWTVSDISKTGVRVKASSSAERIFEAEEIKRCLDLVMGNDEGGMETRRNARIWKDLARETAKESGMSYKNLKAFVDEIAGG
>EVM%20prediction%20scaffold212_size72660.2.mRNA1
MVNLRILLVTFPAQGHINPALQFAKRLVALGAHVTFSTSFGAERMMDKTGASVPGLSFSAFHDGTEHSFRPTDDVDHYMFQLKHCGYNSLAQLIVASAEAGHPFTCVVYSNLIPWVGKVARDFHLPSFLLWNQSATVFDVFYHYFNGFEDAIKMSMSDPAFQLKLPGLSLSFASHDFPSFFSPTNRHTFGLKLMKEQLQILDQETNPPKVLVNTFDSLESEALKSVGRYNLVGVGPLIPSAFLDGKDPSENSFGGDLFRASKDYMIWLDSQPQSSVIYISFGSVSVISKQEKEEMARALLDVGRPFLWVIRTGDAGGSEEKEDKPSCMEELERQGMIVPWCSQVEVLSHPSLGCFVTHCGWNSTLESLTSGVPVVAMPRWTDQLTNARLVNDISRTGVRVKSSSSAEKMFEAEEIKSCLDLVMGNEERGMEVRRNARKWKDLARETAKESGMSYKNLKAFVDEIAGGRDCT
Gene tree
Gene tree

Sheng jun & Dong yang Teams, Yunnan agriculture University